Research
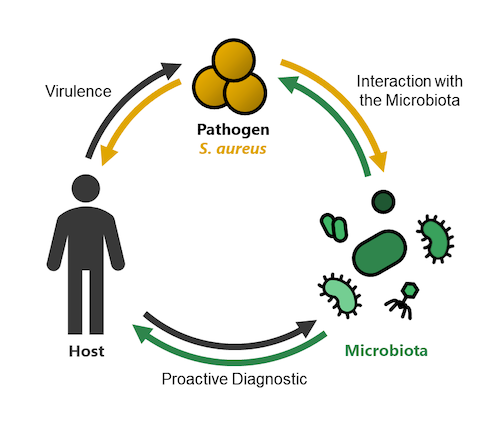
One third of the human population carries the invasive pathogen Staphylococcus aureus within the nasal cavity. This colonization increases the risk of infection and antibiotic resistant lineages (especially MRSA) put urgency on the development of new strategies to prevent and treat infections. The Heilbronner lab addresses this problem in three distinct research topics.
1) Virulence: We investigate the molecular mechanisms of S. aureus pathogenicity during invasive disease with a special focus on trace metal acquisition.
2) Interaction with the Microbiota: We investigate if bacteria of the nasal microbiome can provide resilience against S. aureus colonization.
3) Proactive Diagnostic: Using clinical cohorts, we investigate if metagenome sequencing of nasal specimen can identify microbiome signatures (biomarkers) predicting the risk of a patient to be infected by S. aureus.
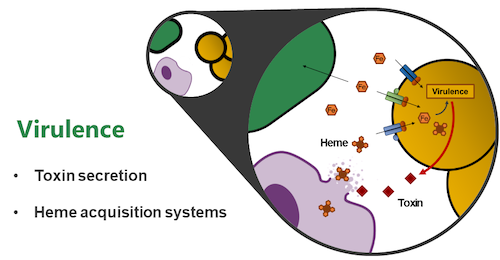
Research Topic 1: Molecular Mechanisms of Staphylococcal Virulence
In order to cause infection, pathogens need to overcome humoral and cellular immune defenses of the host. Additionally, pathogens need to acquire host-derived nutrients and adhere to extracellular matrix molecules.
Our major focus lies on the molecular mechanisms of staphylococcal trace metal acquisition during infection. Iron is essential for bacterial proliferation and the active limitation of iron within the human body is an important immune effector mechanism. However, within humans, iron-containing heme is used for oxygen transport and pathogens have developed complex systems to acquire and degrade heme. Staphylococci secrete cytolytic toxins to release hemoproteins from host cells. In a second step, complex systems employing cell wall-anchored receptors, and membrane ABC-transporters allow coordinated transport of heme across the cell envelope. We study the molecular architecture of these systems on the cellular level using state of the art genetic, biochemical, and in vivo approaches.
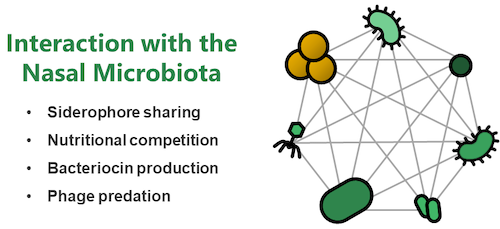
Research Topic 2: Molecular Interactions with the Nasal Microbiome
Nasal colonization of S. aureus represents a major reservoir for infection. However, it is incompletely understood why some human individuals carry S. aureus while others show resilience towards colonization. Within the nasal cavity S. aureus needs to interact with other bacteria of the nasal microbiome and bacterial interactions can be of collaborative or competitive nature. We use extensive strain collections of nasal bacterial isolates and study their interaction with S. aureus. We use high-throughput approaches to identify networks of reciprocal collaboration and support and investigate the underlying molecular mechanisms. Of major focus are interactions that are based on shared nutritional resources (mucins, iron-binding siderophores etc.), bacteriocins (antibacterial molecules) and bacteriophages (viruses infecting bacteria). Hypothesis deriving from our in vitro experiments are tested in mice harboring a humanized nasal microbiome. Our overarching aim is to develop nasal probiotics to displace the pathogen from the nasal cavity of humans.
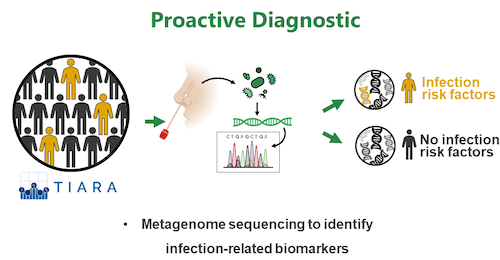
Research Topic 3: Proactive Diagnostics
It is known that S. aureus nasal colonization represents a risk factor for infection with the endogenous strain. However, direct means to translate this knowledge into clinical applications is currently lacking. In frame of the German Centre For Infection Research (DZIF) we use clinical cohorts and full metagenome sequencing of nasal specimen to identify microbiome signatures (e.g. pathogen abundance, presence/absence of specific commensals) that correlate with the development of infection. Such biomarkers are of high relevance as they can encourage proactive treatment of certain patients before infection occurs.
Selected publications (*- Corresponding author)
Original Research Articles
- Zhao Y, Bitzer A, Power JJ, Belikova D, Torres Salazar BO, Adolf LA, Gerlach D, Krismer B, Heilbronner S*. Nasal commensals reduce Staphylococcus aureus proliferation by restricting siderophore availability. ISME J, 18(1). https://doi.org/10.1093/ismejo/wrae123 (2024)
- Sekar S, Schwarzbach S, Nega M, Bloes DA, Smeds E, Kretschmer D, Foster TJ, Heilbronner S*. SLUSH peptides of the PSMβ family enable Staphylococcus lugdunensis to use erythrocytes as a sole source of nutrient iron. The FASEB Journal. 2024;38:e23881. doi:10.1096/fj.202400335R (2024)
- Puls JS, Winnerling B, Power JJ, Krüger AM, Brajtenbach D, Johnson M, Bilici K, Camus L, Fließwasser T, Schneider T, Sahl HG, Ghosal D, Kubitscheck U, Heilbronner S, Grein F. The Staphylococcus epidermidis bacteriocin A37 kills natural competitors with a unique mechanism of action. ISME J, 18(1). https://doi.org/10.1093/ismejo/wrae044 (2024)
- Rosenstein R, Torres Salazar BO, Sauer C, Heilbronner S, Krismer B, Peschel A. The Staphylococcus aureus-antagonizing human nasal commensal Staphylococcus lugdunensis depends on siderophore piracy. Microbiome 12, 213, doi:10.1186/s40168-024-01913-x (2024)
- Adolf LA., Müller-Jochim A., Kricks L., Puls JS., Lopez D., Grein F., Heilbronner S* Functional membrane microdomains and the hydroxamate siderophore transporter ATPase FhuC govern Isd-dependent heme acquisition in Staphylococcus aureus Elife, 12, doi:10.7554/eLife.85304 (2023)
- Krauss S, Harbig TA, Rapp J, Schaefle T, Franz-Wachtel M, Reetz L, Elsherbini AMA, Macek M, Grond S, Link HP, Nieselt K, Krismer B, Peschel A, Heilbronner S* Horizontal transfer of bacteriocin biosynthesis genes requires metabolic adaptation to improve compound production and cellular fitness. Microbiol Spectr, e0317622 (2023)
- Du X, Larsen J, Li Min, Walter A, Both A, Sanchez Carballo PM, Stegger M, Lehmann E, Liu Y, Liu J, Schade J, A.Duda K, Krismer B, Heilbronner S, Weidenmaier C, Slavetinsky C, Mayer M, Rohde H, Winstel, Peschel A. Emerging Staphylococcus epidermidis alters surface glycopolymers to shift from commensal to pathogen behavior. Nat Microbiol 6, 757-768 (2021)
- Belikova D, Jochim A, Powers J, Holden MT, Heilbronner S*. Gene accordions cause genotypic and phenotypic heterogeneity in clonal populations of Staphylococcus aureus. Nat Commun 11, 3526 doi:10.1038/s41467-020-17277-3 (2020).
- Jochim A, Adolf L, Belikova D, Schilling NA, Setyawati I, Chin D, Meyers S, Verhamme P, Heinrichs DE, Slotboom DJ, Heilbronner S*. An ECF-type transporter scavenges heme to overcome iron-limitation in Staphylococcus lugdunensis. Elife 9, doi:10.7554/eLife.57322 (2020).
Reviews
- Adolf LA. and Heilbronner S*, Nutritional Interactions between Bacterial Species Colonising the Human Nasal Cavity: Current Knowledge and Future Prospects. Metabolites 2022 12(6) (2022)
- Heilbronner S*, Brötz-Österheld H, Krismer B, Peschel A. The microbiome-shaping roles of bacteriocins. Nat. Rev. Microbiol. doi:10.1038/s41579-021-00569-w (2021)
- Heilbronner S* and Foster TJ, Staphylococcus lugdunensis. A skin commensal with invasive pathogenic potential. Clin Microbiol Rev, 2021. 34(2). (2021)
Book Chapters
- Torres Salazar B, Lange A. Camus L, Heilbronner S*. Sampling of Human Microbiomes to Screen for Antibiotic-Producing Commensals. Methods Mol Biol 2601, 39-54 (2022)
We are looking for medical doctors interested in the MD-PhD program of the German Center for Infection Research (DZIF)
Additionally, prospective PhD students and Postdocs that are interested in working with us and willing to apply for individual funding (See list below) are welcome to contact Simon Heilbronner
Postdoc funding opportunities:
PhD funding opportunities: